Exercise 7 - Classification II
Task 1
Python exercise for classification based on a multivariate Gaussian classifier.
Step 1: Implement a Gaussian classifier using a d-dimensional feature vector
For the algorithm, see lecture slides or the notes.
It is recommended that you use the built-in functions for computing matrix inverse, determinant, means and covariance matrices, but you should implement the rest of the algorithm yourself.
import numpy as np
# Matrix determinant and inverse
A = np.array([[1, 2, 3], [2, 2, 6], [3, 6, 1]])
det_A = np.linalg.det(A)
# Check if A is invertible
if det_A != 0:
A_inv = np.linalg.inv(A)
# Mean and covariance
## From matrix
B = np.array([[1, 2, 3], [4, 6, 8]])
cov_B = np.cov(B)
col_mean = np.mean(B, axis=0)
row_mean = np.mean(B, axis=1)
## From vectors
c = np.array([1, 2, 3])
d = np.array([4, 6, 8])
cov_cd = np.cov(c, d)
mean_c = np.mean(c)
mean_d = np.mean(d)
## Check for equality
assert (cov_B == cov_cd).all(), "Unequal covariance"
assert (col_mean == np.mean(np.stack([c, d]), axis=0)).all(), "Unequal column mean"
assert (row_mean == np.stack([mean_c, mean_d])).all(), "Unequal row mean"
Step 2: Train the classifier
Train the classifier using the mask tm_train.png (found
here) to estimate the class-specific mean vector
and covariance matrix.
If you want to verify that your code gives the correct classification labels,
check the resulting classification image you get when you classify the entire
image (all 6 wavelength bands tm1.png, …, tm6.png), against the result
tm_classres.mat (which can be found
here).
In this image, each pixel is assigned a class label in .
You can find the test result as a grayscale image tm_classres.png in the
image folder, but if you want to read the .mat
file directly in python, here is how you do it:
import scipy.io
classres_container = scipy.io.loadmat('/path/to/tm_classres.mat')
print(classres_container.keys()) # For info about what the .mat file contains
classres_image = classres_container['klassim']
# Check your result using the classified image "my_classified_image"
num_equal = np.sum(1*(my_classified_image == classres_image))
Step 3: Find the best feature for classification using single features
Run the classification on the multivariate (6-band) input image. Compute the
percentage of correctly classified pixels when using all features, and compare
it to using single features. The classification accuracy should be computed on
the whole image using the test mask tm_test.png.
Also try the simplyfied covariance matrix . Which version gives the highest classification accuracy?
Task 2 - Finding the decision functions for a minimum distance classifier
A classifier that uses diagonal covariance matrices is often called a minimum distance classifier, because a pattern is classified to the class that is closest when distance is computed using Euclidean distance.
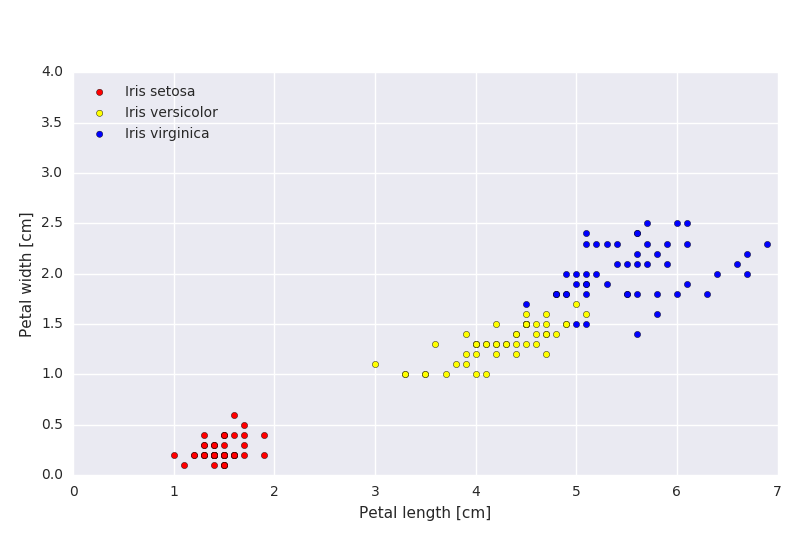
a)
In Figure 2.1, find the class means just by looking at the figure.
b)
If the data is classified using a minimum distance classifier, sketch the decision boundaries on the plot.
Task 3 - Discriminant functions
A classifier that uses Euclidean distance, computes the distance from a point to class as
Show that classification with this rule is equivalent to using the discriminant function
Task 4
In a three-class two-dimensional problem, the feature vectors in each class are normally distributed with covariance matrix
The mean vectors for the three classes are
Assuming that the classes are equally probable (the class prior is uniform):
a)
Classify the feature vector
according to a Bayesian classifier with the given covariance matrix.
b)
Draw the curves of equal Mahalanobis distance from the class with mean
Task 5
Given a two-class classification problem with equal class prior distribution
and Gaussian likelihoods
with means
and covariance matrix
Classify the feature vector
using Bayesian classification.